
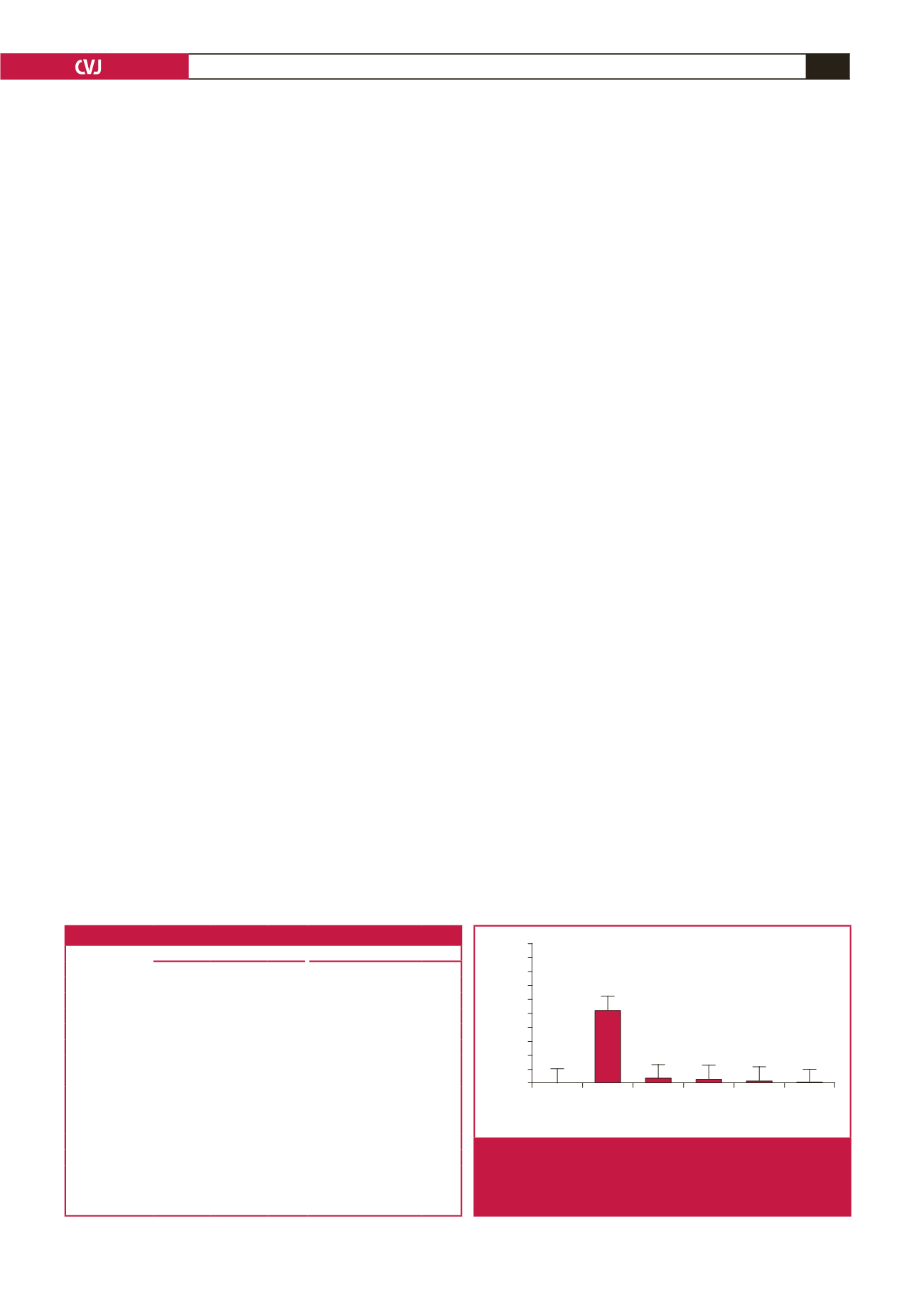
CARDIOVASCULAR JOURNAL OF AFRICA • Volume 27, No 4, July/August 2016
AFRICA
233
were used as dependent variables (
ŷ
). ApoM was used as the
independent variable (
x
). When HDL-C, FPG and TG were
used as three indicators in the regression equation (
x
1
,
x
2
and
x
3
,
respectively), the resulting general multiple regression equation
was
ŷ
=
3.592 + 9.767
x
1
– 0.539
x
2
– 0.464
x
3
with the regression
coefficient test, with
p
=
0.000 and a coefficient of determination
of 0.651. The standardised regression equation was
ŷ
=
0.
573x
1
– 0.301
x
2
– 0.138
x
3
. The closest relationship was between serum
HDL-C and ApoM levels (Table 3).
Association of polymorphisms in the ApoM
proximal promoter region with CAD
DNA sequencing of the polymorphic regions in the proximal
promoter of the ApoM gene revealed that SNPs T-778C and
T-855C of the ApoM gene were valid in the Han Chinese. The
SNPs were accurately detected by PCR-RFLP (Fig. 1). Table 4
shows the plasma lipid and ApoM levels in each group according
to the SNP status.
In the non-CAD group, we found no significant difference in
lipid plasma levels between the TT and the TC+CC groups. In
the CAD group, the TC, TG and ApoM levels were significantly
different between the TT and TC/CC groups. The ApoM plasma
levels in the TT and TC+CC groups were 10.35
±
4.41 and 6.46
±
4.06 μg/ml, respectively in T-778C, and 10.17
±
5.68 and 6.07
±
4.70 μg/ml, respectively in T-855C (both
p
<
0.05; Table 4).
PCR amplification of the human ApoM gene
promoter region
From the above results, that patients with the TC/CC genotype
showed lower plasma ApoM levels compared to those with
the TT genotype, and that ApoM levels were lower in CAD
compared to non-CAD patients, we inferred that the ApoM
promoter variation may alter the promoter activity. To verify
whether the −778T
→
C and −855T
→
C variation affected the
ApoM promoter activity, we applied a reporter gene assay to
detect the luciferase expression with transfection of the specific
allele(s) of the polymorphism.
We designed PCR primers to amplify a 1562-bp slice of the
promoter region of ApoM. Four reporter plasmids, containing
regions −855T to −778T, −855T to –778C, −855C to −778T and
−855C to −778C, were named PGL3-TT, PGL3-TC, PGL3-
CT and PGL3-CC, respectively. The luciferase activities of the
PGL3-TC/CC promoters were lower than those of the PGL3-TT
promoter (PGL-TT, 1.67
±
0.14; PGL-TC, 1.28
±
0.11; PGL-TC,
0.77
±
0.21; PGL-CC, 0.25
±
0.10,
p
=
0.001; Fig. 3)
Transcription factor prediction
The TRANSFAC online prediction software was used to predict
the combined TF binding sites upstream of the ApoM gene (Fig.
4A, B). The binding sites were identified on the ApoM promoter
for some TFs, including Sp1 and AP-1. The AP-2 binding sites
were located at -479 to -488 bp, -349 to -358 bp and -104 to -113
bp on the ApoM promoter. TRANSFAC was used to predict TF
binding sites for the normal and mutated ApoM-855 and ApoM-
778 sites (Fig. 4C–F).
ApoM T-855C provided binding sites for AP-2. ApoM-778T
but not ApoM-778C had binding sites for hepatocyte nuclear
factor-3 (HNF-3), CCAAT enhancer binding protein (C/EBP)
and TATA box-binding protein (TBP). We also used the TESS
online software to predict TF binding sites of the normal and
mutated ApoM-855 (Fig. 4G, H). The results showed that ApoM
T-855C provided binding sites for AP-2.
EMSA results
Small molecular weight chains have faster mobility in EMSA,
whereas the electrophoretic velocity will vary with probe and
protein binding. The velocity of the electrophoretic band
indicates the presence or absence of binding.
We observed a hysteresis band when the C probe but not
the T probe was incubated with nuclear protein (Fig. 5A). The
competitive inhibition test showed that the hysteresis band of
the C probe incubated with nuclear protein could be suppressed
by unlabelled C probe, but not by unlabelled T probe (Fig. 5A).
The supershift test showed that incubating the C probe with
the AP-2
α
antibody produced a more lagging band (Fig. 5B),
whereas the Sp1 antibody did not have this effect (Fig. 5C). The
antibody activity of Sp1 was verified by immunohistochemistry
analyses (Fig. 6A).
Table 4. Lipid profiles according to genotype
CAD
Control
Lipid parameter
TT
CT+CC
p-value
TT
CT+CC
p-value
rs805296
TC (mmol/l)
4.61
±
1.37 4.77
±
1.86 0.030
#
4.33
±
0.49 4.31
±
0.52 0.900
TG (mmol/l)
1.93
±
1.01 3.05
±
2.02 0.002
#
1.03
±
0.82 0.82
±
0.32 0.157
LDL-C (mmol/l)
2.78
±
1.18 2.60
±
1.46 0.610 2.53
±
0.41 2.60
±
0.44 0.655
HDL-C (mmol/l)
1.05
±
0.23 1.01
±
0.34 0.557 1.32
±
0.22 1.35
±
0.19 0.706
ApoM (μg/ml)
10.35
±
4.41 6.46
±
4.06 0.009
#
13.22
±
9.18 3.57
±
3.86 0.007
#
Rs9404941
TC (mmol/l)
4.43
±
1.39 5.30
±
1.49 0.017
#
4.34
±
0.49 4.23
±
0.54 0.542
TG (mmol/l)
1.95
±
1.32 2.70
±
1.04 0.020
#
1.02
±
0.38 0.97
±
0.24 0.711
LDL-C (mmol/l)
2.66
±
1.11 3.03
±
1.52 0.237 2.54
±
0.40 2.46
±
0.50 0.604
HDL-C (mmol/l)
1.07
±
0.26 0.97
±
0.20 0.123 1.32
±
0.21 1.32
±
0.26 0.990
ApoM (μg/ml)
10.17
±
5.68 6.07
±
4.70 0.009
#
12.87
±
9.40 8.30
±
6.45 0.184
Data are means
±
SD.
#
Statistical significance (
p
<
0.05) for TC/CC genotype group vs TT group
with Dunnett’s test.
TC, total cholesterol; TG, triglyceride; LDL-C, HDL-C, low- and high-density lipoprotein choles-
terol, respectively; ApoM, apolipoprotein M.
PGL3-
basic
PGL3-
control
group
PGL3TT PGL3TC PGL3CT PGL3CC
Influence of SNPs T-778C and T-855C on ApoM
prometer activity with liver cell extracts
ApoM promoter activity
50
45
40
35
30
25
20
15
10
5
0
Fig. 3.
Relative luciferase activity of the ApoM promoter
for each transfected group. Data are means
±
SD.
All groups were significantly different (
p
<
0.05 by
Dunnett’s test) compared to the PGL3-TT group.

















