
CARDIOVASCULAR JOURNAL OF AFRICA • Volume 27, No 4, July/August 2016
234
AFRICA
Luciferase activity detection with AP-2
α
overexpression or interference
To examine the role of the TF AP-2
α
in mediating ApoM
promoter activity, we treated HepG2 cells expressing the mutant-
type or wild-type allele of the ApoM-855 site with siRNAs
against AP-2
α
(Fig. 7A, B) or with an AP-2
α
expression vector
(Fig. 7C–F), and examined changes in the luciferase activity of
the -855 site (Fig. 8).
With AP-2
α
interference, the luciferase activity in cells
expressing the wild-type allele (1.63) was less elevated compared
to cells expressing the mutant-type allele (2.99). The luciferase
activities of treated cells were increased compared to the
1
2
3
4
5
DNA–protein
complex
free probe
free probe
AP-2
α
DNA–protein
complex
mutant probe
+
nuclear protein
+
Sp1 antibody
mutant probe
+
nuclear protein
Fig. 5.
Experimental results for transcription factor binding in the ApoM promoter region. A. EMSA. Lane 1: biotin-labelled T probe
+ nuclear protein (NE); lane 2: biotin-labelled C probe; lane 3: biotin-labelled C probe + NE; lane 4: biotin-labelled C probe +
NE + unlabelled C probe; and lane 5: biotin-labelled C probe + NE + unlabelled T probe. B. AP-2
α
supershift. Biotin-labelled
C probe + NE + AP-2
α
antibody. C. Sp1 supershift. Biotin-labelled C probe + NE + Sp1 antibody.
A
B
C
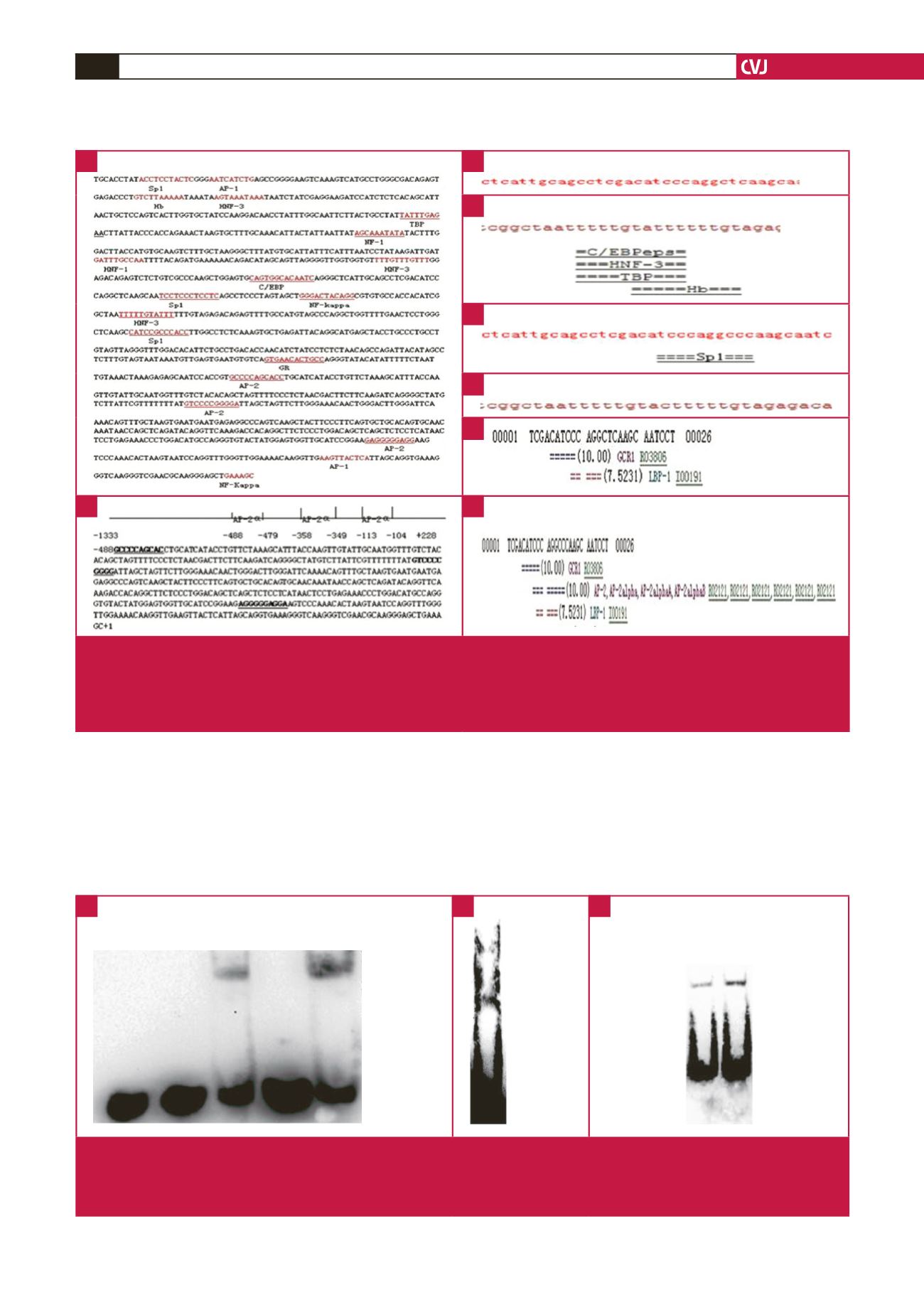
Fig. 4.
Prediction of transcription factor binding in the ApoM promoter region. A. Transcription factor binding prediction for ApoM.
B. AP-2
α
potential binding sites of ApoM start promoter sequence. Mutation site analysis for ApoM-855 and ApoM-778 by
TRANSFAC (C–F), and by TESS (G–H). Binding sites with wild-type alleles of ApoM-855 (C), and ApoM-778 (D), and bind-
ing sites with mutant-type alleles of ApoM-855 (E), and ApoM-778 (F) by TRANSFAC. Binding sites with wild-type (C), and
mutant-type alleles (D) of ApoM-855 by TESS.
A
B
C
D
E
F
G
H

















