
CARDIOVASCULAR JOURNAL OF AFRICA • Volume 27, No 3, May/June 2016
AFRICA
155
and rs117939448). Two novel single-nucleotide polymorphisms
were found in
MYH7
(c.1368C
>
T) and
PRKAG2
(c.828C
>
A) in
two different probands.
The total number of variants in the 15 genes per HCM
patients (regardless of whether it was disease causing or not)
ranged from six to 20 (mean: 12.8, SD 3.2; median 12). The
patient who died had a higher number of variants (14.8
±
3.9)
compared to survivors (12.5
±
3.1), although this difference was
not statistically significant (
p
=
0.47).
Outcome of HCM in South Africans
The mean duration of follow up was 9.1
±
3.4 years. Of the 43
patients studied, eight died during the period of follow up. The
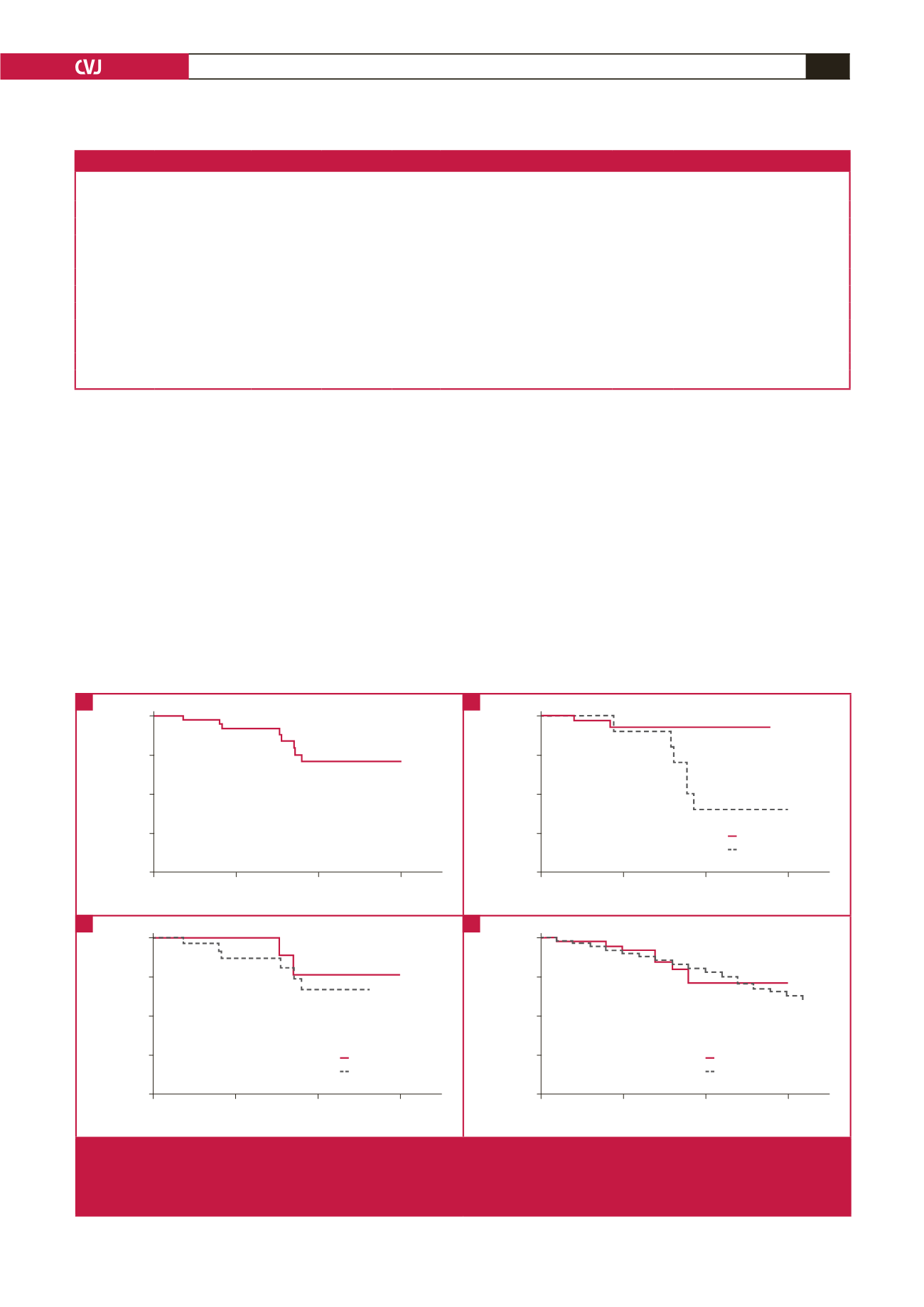
overall Kaplan–Meier survival estimate is shown in Fig. 1A; the
cumulative proportion of patients who survived to 10 years was
74%. Complications of chronic heart failure, atrial fibrillation,
stroke and evolution to dilated cardiomyopathy with systolic
dysfunction were observed in 11 (25.6%), eight (18.6%), four
(9.3%) and four (9.3%), respectively.
Therapeutic interventions, including surgical myomectomy,
alcohol septal ablation and orthotopic heart transplantation
were performed on three (7.0%), one (2.3%) and one (2.3%)
patients, respectively. At the last visit, 12 (27.9%) reported
NYHA functional class III and IV performance status (Table
4). The most frequently prescribed drugs were beta-blockers and
calcium channel blockers, used by 33 (76.7%) and 17 (39.5%)
patients, respectively (Table 5).
Table 3. Disease-causing mutations found in 10 unrelated index cases with hypertrophic cardiomyopathy
Index case ID Ethnicity
Reported
previously?
Gene
Exon Nucleotide and amino acid change
Type of
mutation
Reference
HCM1.1
Indian
Yes
MYH7
5
c.611G
>
A (p.R204H)
Missense
Richard,
et al
. 2003
18
HCM4.1
Mixed ancestry
No
MYH7
20
c.2282C
>
A (p.T761N)
Missense
Novel
HCM7.1
Mixed ancestry
Yes
MYH7
31
c.4258C
>
T (p.R1420W)
Missense
Zou,
et al.
2013
22
HCM11.1
Mixed ancestry
Yes
MYBPC3
12
c.1000G
>
A (p.E334K)
Missense
Bahrudin,
et al
. 2008
23
HCM14.1
European
No
MYH7
20
c.2167C
>
T (p.R723C)
Missense
Novel
HCM16.1
European
Yes
MYH7
9
c.746G
>
A (p.R249Q)
Missense
Zou,
et al.
2013
22
HCM21.1
Black African
Yes
MYBPC3
6
c.772G
>
A (p.E258K)
Missense
Andersen,
et al
. 2004
24
HCM33.1
Mixed ancestry
Yes
MYBPC3
5
c.530G
>
A (p.R177H)
Missense University of Stellenbosch thesis
25
HCM34.1
Black African
Yes
MYH7
14
c.1357C
>
T (p.R453C)
Missense
Zou,
et al
. 2013
22
HCM38.1
Black African
Yes
MYBPC3
15
c.1246G
>
A (p.G416S)
Missense
Tanjore,
et al.
2008
26
HCM, hypertrophic cardiomyopathy;
MYBPC3
, cardiac myosin-binding protein C;
MYH7
, cardiac
β
-myosin heavy chain.
0
5
10
15
Survival time (years)
Cumulative proportion surviving
1.00
0.75
0.50
0.25
0.00
0
5
10
15
Survival time (years)
Cumulative proportion surviving
1.00
0.75
0.50
0.25
0.00
NYHA I, II
NYHA III, IV
p
=
0.026
0
5
10
15
Survival time (years)
Cumulative proportion surviving
1.00
0.75
0.50
0.25
0.00
Mutation identified
No mutation
p
=
0.474
0
5
10
15
Survival time (years)
Cumulative proportion surviving
1.00
0.75
0.50
0.25
0.00
Cohort survival
SA population survival
p
=
0.531
Fig. 1.
Kaplan–Meier survival estimates in HCM. A. Kaplan–Meier plot showing the survival of HCM patients; B. Kaplan–Meier plot
showing the survival of HCM patients when stratified by NYHA functional class; C. Kaplan–Meier plot showing the survival
of HCM patients when stratified by the presence or absence of HCM-causing mutation(s); D. Kaplan–Meier plot showing
the survival of HCM patients when compared to age-, gender- and race-matched members of the South African population.
A
C
B
D

















